QSHGM, a database of QS in Human Gut Microbiota includes 28,567 redundancy removal QS synthases (1,882) and receptors (26,685) entries for 818 gut microbes that are mainly Firmicutes (79), Actinobacteria (36), Proteobacteria (69), Bacteroidetes (16) and others (10).
We have collected and sorted nine common QS signal synthases (AHLs, CAI-1, HAQs, DSFs, Dialkylresorcinols, Photopyrones, AIPs, Indole, and AI-2) for 210 microbial genus as the heatmap representation listed in Figure 1. AHLs are only found in Proteobacteria, AIPs are mostly existed in Firmicutes, and other QS signals are distributed in-homogeneously in the whole genus-level microbes. Among the 818 human gut microbes, Proteobacteria and Bacteroidetes have denser QS signals distributions than Firmicutes and Actinobacteria.
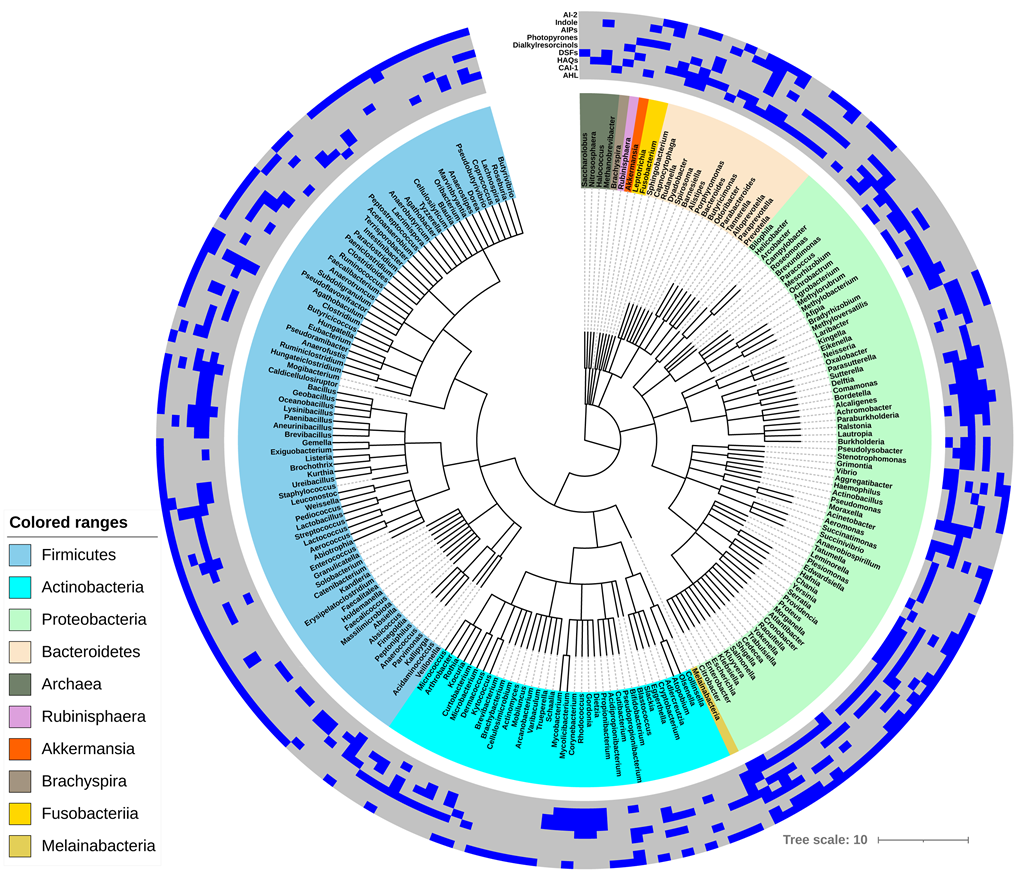
Figure 1. Hierarchical clustering of nine QS signals found in 818 human gut microbes. The heatmap on the outermost layer indicates the QS signals distribution in each cluster, existence is colored in blue; no existence, gray.
To understand the microbial language (QS signals) among different genus, we have also analyzed the genus distributions based on nine languages. Take six languages (AHLs, CAI-1, HAQs, DSFs, Indole, and AI-2) as example, we found that there are 64, 40, 22 and 5 overlaps for two, three, four, and five QS languages, respectively (Figure 2).
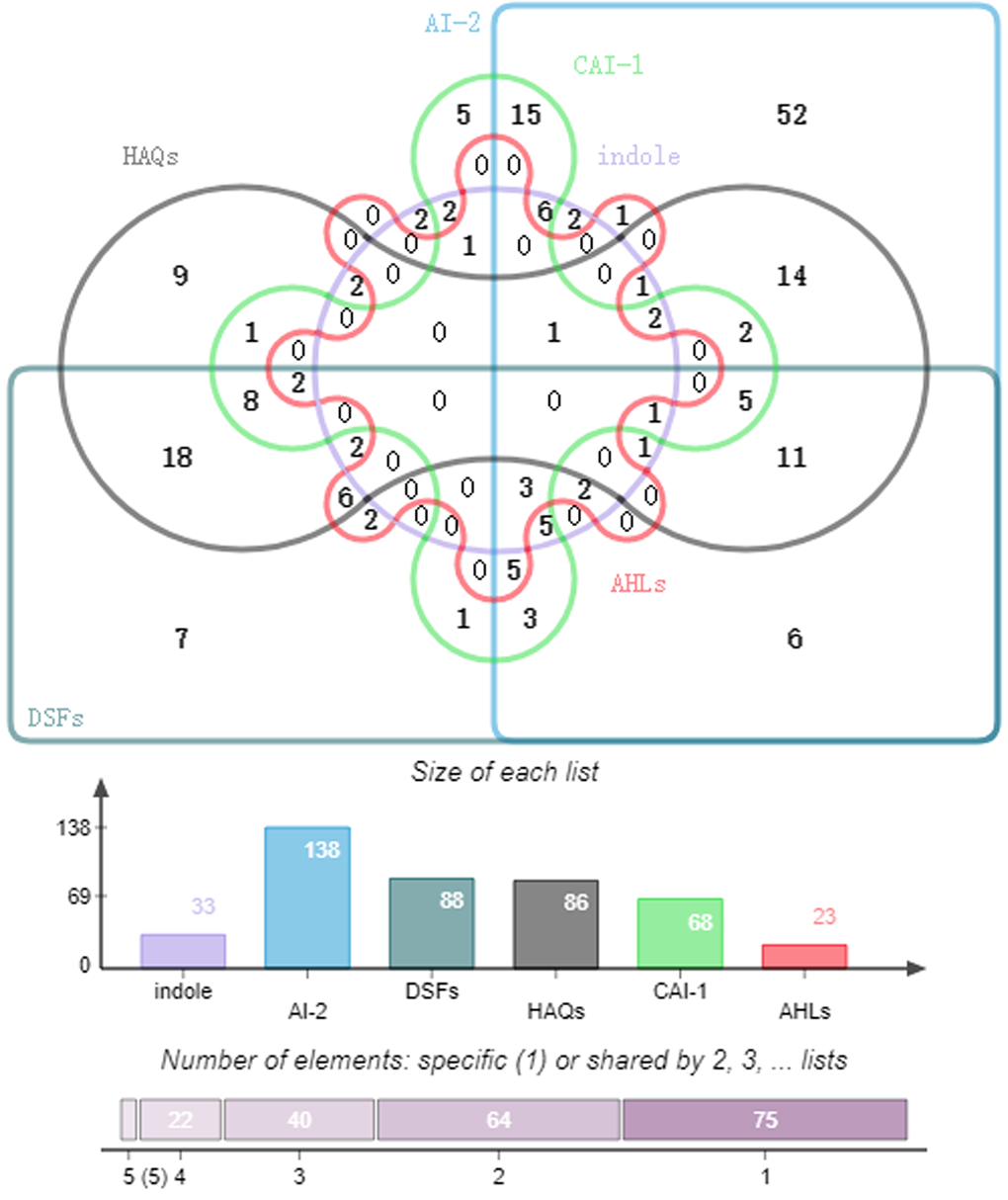
Figure 2. Microbial genus distribution based on six QS signals.
As stated above, microbes could communicate via various QS signals, so it would be possible to construct cell-cell communication networks among different bacteria based on diverse languages, which can be termed as QS social network. We have constructed a genus-level communication network based on above nine QS languages (Figure 3). It intricates network visualizes the complex communications and interactions among human gut microbiota.
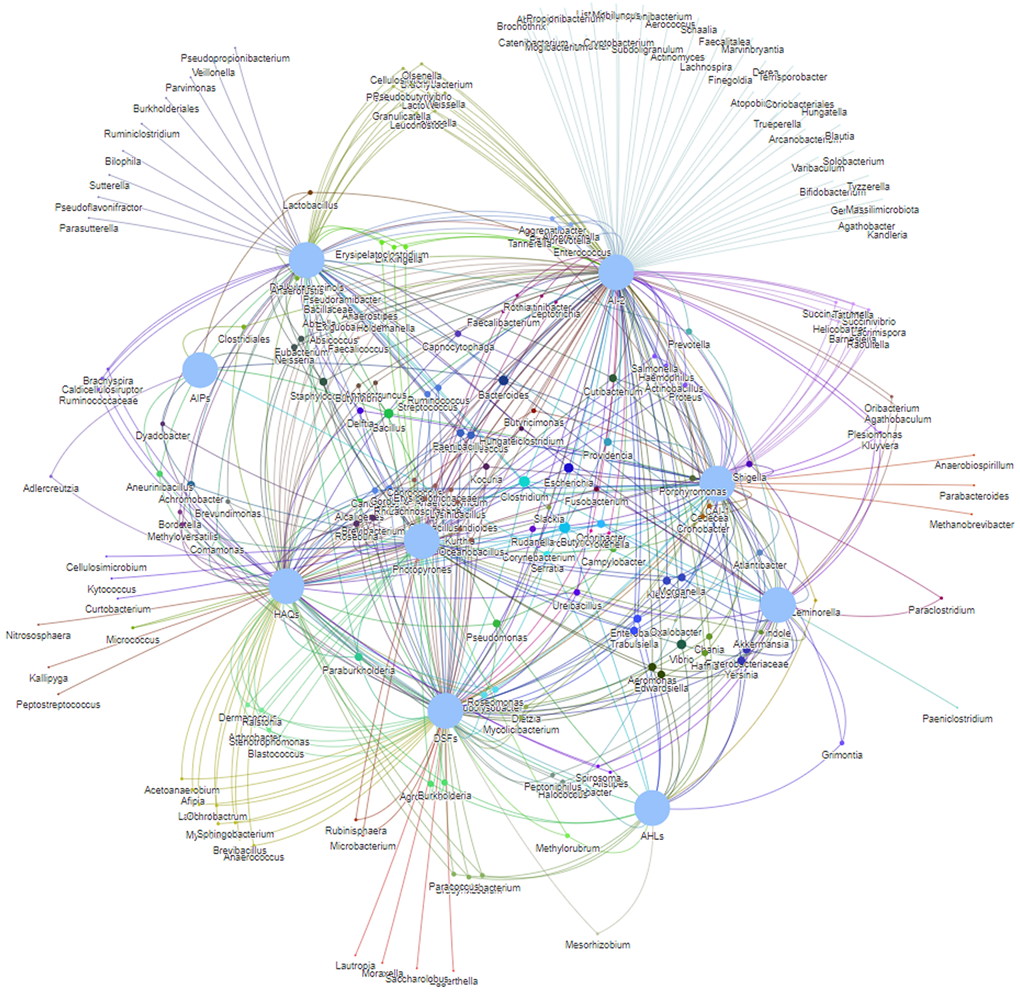
Figure 3. The QS communication network based on nine QS signals.
Take the seven-strain simplified human microbiomes (SIHUMIs) used by Colosimo et al as an example, we constructed a QS social network that includes QS signals producing and receiving (Figure 4). TCS, as a common and typical QS system, widely exists in most microbes. QS signal reception is more varied than signal production, which means that some microbes can receive a particular signal without producing it. This phenomenon is consistent with the cheating behavior in many microbial systems.
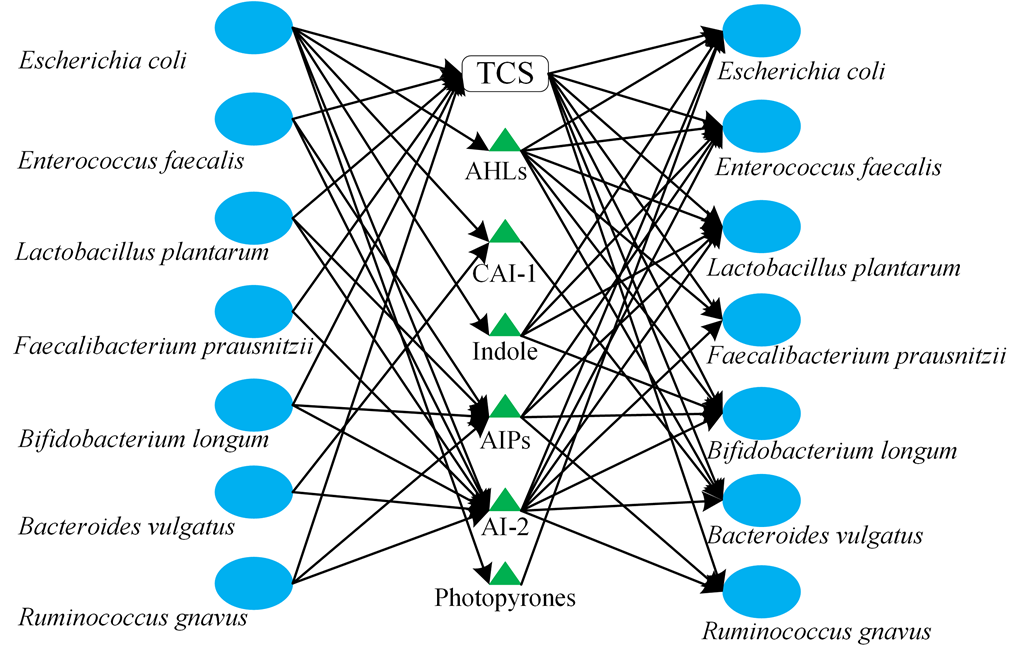
Figure 4. A simplified QS communication network that includes QS signals producing and receiving.
Although QS complexity for multi-layer control, bio-molecular variability, and QS crosstalk for microbiota, there is huge potential to construct a comprehensive QS social network including signals sending and receiving for human gut microbiota based on the QS synthases and receptors. On the one hand, the comprehensive QS social network can be used to analyze microbial communication principles, make the dynamics and resilience of the complex microbial communities more predictable. On the other hand, the ability to engineer microbes or QS signals levels in diverse QS social networks may provide opportunities for future anti-infective strategies, reducing the antibiotics resistance, and developing potential therapies for various diseases.